Synthetic biology and biomanufacturing are buzzwords of an emerging bioeconomy based on our increasing ability to manipulate living systems. This workshop will equip bioscience educators with the knowledge, skills, and resources to introduce students to a simple workflow to manufacture bacteriophages (phages) with new host specificity, illustrating how biomolecules are adapted to interact with cell-surface receptors.
Join Vincent Noireaux, an instructor of the annual Synthetic Biology course at Cold Spring Harbor Laboratory, to come up-to-the minute with a new, cell-free transcription-translation (TXTL) system he developed to biomanufacture phages in vitro. This workshop emerges from a distinguished lineage of bacteriophage research that began at Cold Spring Harbor Laboratory (CSHL) and provided the first tools to explore the molecular mechanics of living cells. The “Phage Course,” founded at CSHL in 1945 by Max Delbrück and Salvador Luria, trained the first two generations of molecular biologists. Al Hershey and Martha Chase’s “blender experiment,” conducted at CSHL in 1952, provided conclusive evidence that DNA is the molecule of heredity. Delbrück, Luria, Hershey shared the 1969 Nobel Prize for this seminal work.
Bacteriophages are being rediscovered as powerful tools to meet the pharmaceutical challenge of untreatable, multi-drug resistant bacterial infections. In the food industry, bacteriophages prevent formation of biofilms on equipment surfaces, sanitize fresh fruits and vegetables, and extend the shelf life of packaged foods. Using phages to treat bacterial infections in livestock increases resilience and helps eliminate the reservoir of antibiotic-resistant bacteria. Phages also provide a virtually limitless source of bioactive materials that are increasingly exploited in biotechnology, nanotechnology, and bioremediation.
Phages are used in education as examples of simple genetic systems. The SEA-PHAGES Program of the Howard Hughes Medical Institute annotates phage genomes, and is one of the most widely implemented infrastructures for course-based undergraduate research experiences (CUREs). TXTL is a logical next step for students who have been exposed to phage and/or bacterial genetics, providing them an opportunity to explore the use of phages in biomanufacturing.
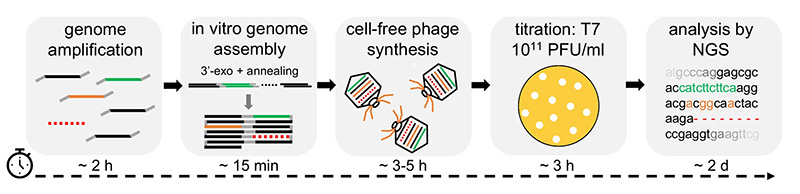
Workshop participants will conduct hands-on experiments to express reporter genes and whole phage genomes in vitro, using a cell-free extract. Participants will complete an entire workflow to engineer wild-type T7 phage to infect a new E. coli host. This begins with long PCRs to amplify several fragments of the T7 genome, plus PCR mutagenesis of the tail fiber gene. The PCR products are then assembled and packaged as complete T7 genomes using the cell-free TXTL system. Spotting assays compare the host range of wild-type and mutant T7 phages. Participants will then conduct nanopore DNA sequencing, delivering same-day results to investigate point mutations in the tail fiber gene that account for infectivity.
A $500 stipend will be provided. Travel funds are available.